- Record: found
- Abstract: found
- Article: found
Implications of TGFβ on Transcriptome and Cellular Biofunctions of Palatal Mesenchyme
Read this article at
Abstract
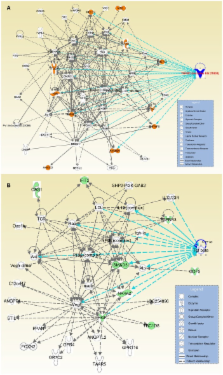
Development of the palate comprises sequential stages of growth, elevation, and fusion of the palatal shelves. The mesenchymal component of palates plays a major role in early phases of palatogenesis, such as growth and elevation. Failure in these steps may result in cleft palate, the second most common birth defect in the world. These early stages of palatogenesis require precise and chronological orchestration of key physiological processes, such as growth, proliferation, differentiation, migration, and apoptosis. There is compelling evidence for the vital role of TGFβ-mediated regulation of palate development. We hypothesized that the isoforms of TGFβ regulate different cellular biofunctions of the palatal mesenchyme to various extents. Human embryonic palatal mesenchyme (HEPM) cells were treated with TGFβ1, β2, and β3 for microarray-based gene expression studies in order to identify the roles of TGFβ in the transcriptome of the palatal mesenchyme. Following normalization and modeling of 28,869 human genes, 566 transcripts were detected as differentially expressed in TGFβ-treated HEPM cells. Out of these altered transcripts, 234 of them were clustered in cellular biofunctions, including growth and proliferation, development, morphology, movement, cell cycle, and apoptosis. Biological interpretation and network analysis of the genes active in cellular biofunctions were performed using IPA. Among the differentially expressed genes, 11 of them are known to be crucial for palatogenesis ( EDN1, INHBA, LHX8, PDGFC, PIGA, RUNX1, SNAI1, SMAD3, TGFβ 1, TGFβ2, and TGFβ R1). These genes were used for a merged interaction network with cellular behaviors. Overall, we have determined that more than 2% of human transcripts were differentially expressed in response to TGFβ treatment in HEPM cells. Our results suggest that both TGFβ1 and TGFβ2 orchestrate major cellular biofunctions within the palatal mesenchyme in vitro by regulating expression of 234 genes.
Related collections
Most cited references79
- Record: found
- Abstract: found
- Article: not found
Epithelial-Mesenchymal Transition in Cancer: Parallels Between Normal Development and Tumor Progression
- Record: found
- Abstract: found
- Article: not found
"Stemness": transcriptional profiling of embryonic and adult stem cells.
- Record: found
- Abstract: found
- Article: not found