- Record: found
- Abstract: found
- Article: found
An improved clear cell renal cell carcinoma stage prediction model based on gene sets
Read this article at
Abstract
Background
Clear cell renal cell carcinoma (ccRCC) is the most common subtype of renal cell carcinoma and accounts for cancer-related deaths. Survival rates are very low when the tumor is discovered in the late-stage. Thus, developing an efficient strategy to stratify patients by the stage of the cancer and inner mechanisms that drive the development and progression of cancers is critical in early prevention and treatment.
Results
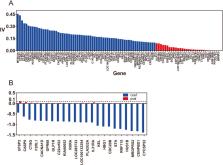
In this study, we developed new strategies to extract important gene features and trained machine learning-based classifiers to predict stages of ccRCC samples. The novelty of our approach is that (i) We improved the feature preprocessing procedure by binning and coding, and increased the stability of data and robustness of the classification model. (ii) We proposed a joint gene selection algorithm by combining the Fast-Correlation-Based Filter (FCBF) search with the information value, the linear correlation coefficient, and variance inflation factor, and removed irrelevant/redundant features. Then the logistic regression-based feature selection method was used to determine influencing factors. (iii) Classification models were developed using machine learning algorithms. This method is evaluated on RNA expression value of clear cell renal cell carcinoma derived from The Cancer Genome Atlas (TCGA). The results showed that the result on the testing set (accuracy of 81.15% and AUC 0.86) outperformed state-of-the-art models (accuracy of 72.64% and AUC 0.81) and a gene set FJL-set was developed, which contained 23 genes, far less than 64. Furthermore, a gene function analysis was used to explore molecular mechanisms that might affect cancer development.
Related collections
Most cited references16
- Record: found
- Abstract: found
- Article: not found
Somatic and germline CACNA1D calcium channel mutations in aldosterone-producing adenomas and primary aldosteronism
- Record: found
- Abstract: found
- Article: not found
Multi-institutional validation of a new renal cancer-specific survival nomogram.
- Record: found
- Abstract: found
- Article: not found