- Record: found
- Abstract: found
- Article: found
Ecological Analyses of Mycobacteria in Showerhead Biofilms and Their Relevance to Human Health
Read this article at
Abstract
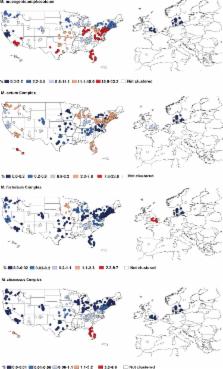
Bacteria thrive in showerheads and throughout household water distribution systems. While most of these bacteria are innocuous, some are potential pathogens, including members of the genus Mycobacterium that can cause nontuberculous mycobacterial (NTM) lung infection, an increasing threat to public health. We found that showerheads in households across the United States and Europe often harbor abundant mycobacterial communities that vary in composition depending on geographic location, water chemistry, and water source, with households receiving water treated with chlorine disinfectants having particularly high abundances of certain mycobacteria. The regions in the United States where NTM lung infections are most common were the same regions where pathogenic mycobacteria were most prevalent in showerheads, highlighting the important role of showerheads in the transmission of NTM infections.
ABSTRACT
Bacteria within the genus Mycobacterium can be abundant in showerheads, and the inhalation of aerosolized mycobacteria while showering has been implicated as a mode of transmission in nontuberculous mycobacterial (NTM) lung infections. Despite their importance, the diversity, distributions, and environmental predictors of showerhead-associated mycobacteria remain largely unresolved. To address these knowledge gaps, we worked with citizen scientists to collect showerhead biofilm samples and associated water chemistry data from 656 households located across the United States and Europe. Our cultivation-independent analyses revealed that the genus Mycobacterium was consistently the most abundant genus of bacteria detected in residential showerheads, and yet mycobacterial diversity and abundances were highly variable. Mycobacteria were far more abundant, on average, in showerheads receiving municipal water than in those receiving well water and in U.S. households than in European households, patterns that are likely driven by differences in the use of chlorine disinfectants. Moreover, we found that water source, water chemistry, and household location also influenced the prevalence of specific mycobacterial lineages detected in showerheads. We identified geographic regions within the United States where showerheads have particularly high abundances of potentially pathogenic lineages of mycobacteria, and these “hot spots” generally overlapped those regions where NTM lung disease is most prevalent. Together, these results emphasize the public health relevance of mycobacteria in showerhead biofilms. They further demonstrate that mycobacterial distributions in showerhead biofilms are often predictable from household location and water chemistry, knowledge that advances our understanding of NTM transmission dynamics and the development of strategies to reduce exposures to these emerging pathogens.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: not found
Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex.
- Record: found
- Abstract: found
- Article: not found
ppcor: An R Package for a Fast Calculation to Semi-partial Correlation Coefficients.
- Record: found
- Abstract: found
- Article: not found