- Record: found
- Abstract: found
- Article: found
Genetic variants and their association with phenotypic resistance to bedaquiline in Mycobacterium tuberculosis: a systematic review and individual isolate data analysis

Read this article at
Summary
Background
Bedaquiline is a crucial drug for control of rifampicin-resistant tuberculosis. Molecular drug resistance assays could facilitate effective use of bedaquiline and surveillance of drug resistance emergence. To facilitate molecular assay development, we aimed to identify genomic markers of bedaquiline resistance.
Methods
In this systematic review and individual isolate analysis, we searched Europe PubMed Central and Scopus for studies published from the inception of each database until Oct 19, 2020, that assessed genotypic and phenotypic bedaquiline resistance in clinical or non-clinical Mycobacterium tuberculosis isolates. All studies reporting on the assessment of variants in the four genes of interest ( Rv0678, atpE, pepQ, and Rv1979c) and phenotypic bedaquiline data in both clinical and non-clinical samples were included. We collated individual isolate data from eligible studies to assess the association between genomic variants with phenotypic bedaquiline resistance, using a standardised method endorsed by WHO. Risk of bias of the extracted data was independently assessed by two authors using the Quality Assessment of Diagnostic Accuracy Studies tool for clinical studies and Systematic Review Center for Laboratory Animal Experimentation tool for animal studies. The primary outcome was to identify mutations associated with resistance in four genes of interest ( Rv0678, atpE, pepQ, and Rv1979c); for each genomic variant, the odds ratio (OR), 95% CI, and p value were calculated to identify resistance markers associated with bedaquiline resistance. This study is registered with PROSPERO, CRD42020221498.
Findings
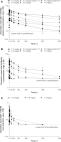
Of 1367 studies identified, 41 published between 2007 and 2020 were eligible for inclusion. We extracted data on 1708 isolates: 1569 (91·9%) clinical isolates and 139 (8·1%) non-clinical isolates. We identified 237 unique variants in Rv0678, 14 in atpE, 28 in pepQ, and 11 in Rv1979c. Most clinical isolates with a single variant reported in Rv0678 (229 [79%] of 287 variants), atpE (14 [88%] of 16 variants), pepQ (32 [100%] of 32 variants), or Rv1979c (115 [98%] of 119 variants) were phenotypically susceptible to bedaquiline. Except for the atpE 187G→C (OR ∞, [95% CI 13·28–∞]; p<0·0001) and Rv0678 138_139insG (OR 6·91 [95% CI 1·16–47·38]; p=0·016) variants, phenotypic–genotypic associations were not significant (p≥0·05) for any single variant in Rv0678, atpE, pepQ, and Rv1979c.
Interpretation
Absence of clear genotypic–phenotypic associations for bedaquiline complicates the development of molecular drug susceptibility tests. A concerted global effort is urgently needed to assess the genotypic and phenotypic drug susceptibility of M tuberculosis isolates, especially in patients who have received unsuccessful bedaquiline-containing regimens. Treatment regimens should be designed to prevent emergence of bedaquiline resistance and phenotypic drug susceptibility tests should be used to guide and monitor treatment.
Related collections
Most cited references59
- Record: found
- Abstract: found
- Article: not found
QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies.
- Record: found
- Abstract: found
- Article: found
SYRCLE’s risk of bias tool for animal studies
- Record: found
- Abstract: found
- Article: not found