- Record: found
- Abstract: found
- Article: found
Dysbiosis of Gram-negative gut microbiota and the associated serum lipopolysaccharide exacerbates inflammation in type 2 diabetic patients with chronic kidney disease
Read this article at
Abstract
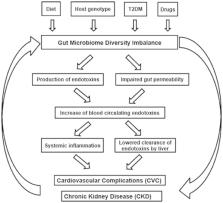
Lipopolysaccharide (LPS), a potent endotoxin present in the outer membrane of Gram-negative bacteria, causes chronic immune responses associated with inflammation. In the present study, the association between LPS and the dysbiosis of Gram-negative bacteria in the gut microbiome was determined in patients with type 2 diabetes mellitus (T2DM) and chronic kidney disease (T2DM-CKD; stages 4 and 5, not on dialysis) compared with healthy individuals. Microbiome diversity was analyzed in patients with T2DM-CKD and healthy controls by sequencing the hypervariable sub-regions of the 16S ribosomal RNA gene from stool samples. Serum samples were assayed by ELISA for LPS, C-reactive protein (CRP), tumor necrosis factor-α (TNFα), interleukin-6 (IL6) and endothelin-1. A total of four gut Gram-negative phyla (Bacteroidetes, Proteobacteria, Fusobacteria and Verrucomicrobia) were identified in the gut microbiome of the T2DM-CKD and control groups. Proteobacteria, Verrucomicrobia and Fusobacteria exhibited significantly increased relative abundance in patients with T2DM-CKD when compared with controls (P<0.05). The levels of serum LPS were significantly increased in patients with T2DM-CKD compared with controls (P<0.05). Elevated serum LPS was significantly correlated with increased levels of TNFα, IL6 and CRP. The dysbiosis of Gram-negative bacteria in the gut microbiome of patients with T2DM-CKD may contribute to the elevated serum levels of LPS and the consequential effects on the inflammatory biomarkers in these patients. The association between the dysbiosis of Gram-negative bacteria in the gut microbiome of patients with T2DM-CKD, increased LPS levels and the effects on inflammatory biomarkers may provide insight into potential diagnostic and therapeutic approaches in the treatment of T2DM-CKD.
Related collections
Most cited references40

- Record: found
- Abstract: found
- Article: not found
Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome.

- Record: found
- Abstract: found
- Article: found