- Record: found
- Abstract: found
- Article: found
Long-Term Evolution of Burkholderia multivorans during a Chronic Cystic Fibrosis Infection Reveals Shifting Forces of Selection
Read this article at
Abstract
Bacteria may become genetically and phenotypically diverse during long-term colonization of cystic fibrosis (CF) patient lungs, yet our understanding of within-host evolutionary processes during these infections is lacking. Here we combined current genome sequencing technologies and detailed phenotypic profiling of the opportunistic pathogen Burkholderia multivorans using sequential isolates sampled from a CF patient over 20 years. The evolutionary history of these isolates highlighted bacterial genes and pathways that were likely subject to strong selection within the host and were associated with altered phenotypes, such as biofilm production, motility, and antimicrobial resistance. Importantly, multiple lineages coexisted for years or even decades within the infection, and the period of diversification within the dominant lineage was associated with deterioration of the patient’s lung function. Identifying traits under strong selection during chronic infection not only sheds new light onto Burkholderia evolution but also sets the stage for tailored therapeutics targeting the prevailing lineages associated with disease progression.
ABSTRACT
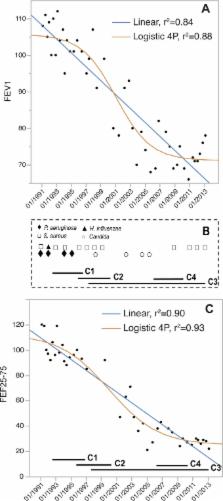
Burkholderia multivorans is an opportunistic pathogen capable of causing severe disease in patients with cystic fibrosis (CF). Patients may be chronically infected for years, during which the bacterial population evolves in response to unknown forces. Here we analyze the genomic and functional evolution of a B. multivorans infection that was sequentially sampled from a CF patient over 20 years. The population diversified into at least four primary, coexisting clades with distinct evolutionary dynamics. The average substitution rate was only 2.4 mutations/year, but notably, some lineages evolved more slowly, whereas one diversified more rapidly by mostly nonsynonymous mutations. Ten loci, mostly involved in gene expression regulation and lipid metabolism, acquired three or more independent mutations and define likely targets of selection. Further, a broad range of phenotypes changed in association with the evolved mutations; they included antimicrobial resistance, biofilm regulation, and the presentation of lipopolysaccharide O-antigen repeats, which was directly caused by evolved mutations. Additionally, early isolates acquired mutations in genes involved in cyclic di-GMP (c-di-GMP) metabolism that associated with increased c-di-GMP intracellular levels. Accordingly, these isolates showed lower motility and increased biofilm formation and adhesion to CFBE41o − epithelial cells than the initial isolate, and each of these phenotypes is an important trait for bacterial persistence. The timing of the emergence of this clade of more adherent genotypes correlated with the period of greatest decline in the patient’s lung function. All together, our observations suggest that selection on B. multivorans populations during long-term colonization of CF patient lungs either directly or indirectly targets adherence, metabolism, and changes in the cell envelope related to adaptation to the biofilm lifestyle.
IMPORTANCE Bacteria may become genetically and phenotypically diverse during long-term colonization of cystic fibrosis (CF) patient lungs, yet our understanding of within-host evolutionary processes during these infections is lacking. Here we combined current genome sequencing technologies and detailed phenotypic profiling of the opportunistic pathogen Burkholderia multivorans using sequential isolates sampled from a CF patient over 20 years. The evolutionary history of these isolates highlighted bacterial genes and pathways that were likely subject to strong selection within the host and were associated with altered phenotypes, such as biofilm production, motility, and antimicrobial resistance. Importantly, multiple lineages coexisted for years or even decades within the infection, and the period of diversification within the dominant lineage was associated with deterioration of the patient’s lung function. Identifying traits under strong selection during chronic infection not only sheds new light onto Burkholderia evolution but also sets the stage for tailored therapeutics targeting the prevailing lineages associated with disease progression.
Related collections
Most cited references83

- Record: found
- Abstract: found
- Article: found
The Sequence Alignment/Map format and SAMtools
- Record: found
- Abstract: found
- Article: not found
Fast gapped-read alignment with Bowtie 2.
- Record: found
- Abstract: found
- Article: not found