- Record: found
- Abstract: found
- Article: found
APOBEC3B can impair genomic stability by inducing base substitutions in genomic DNA in human cells
Read this article at
Abstract
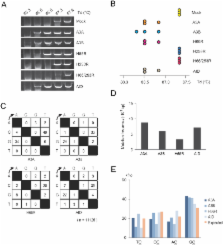
Human APOBEC3 proteins play pivotal roles in intracellular defense against viral infection by catalyzing deamination of cytidine residues, leading to base substitutions in viral DNA. Activation-induced cytidine deaminase (AID), another member of the APOBEC family, is capable of editing immunoglobulin (Ig) and non-Ig genes, and aberrant expression of AID leads to tumorigenesis. However, it remains unclear whether APOBEC3 (A3) proteins affect stability of human genome. Here we demonstrate that both A3A and A3B can induce base substitutions into human genome as AID can. A3B is highly expressed in several lymphoma cells and somatic mutations occur in some oncogenes of the cells highly expressing A3B. Furthermore, transfection of A3B gene into lymphoma cells induces base substitutions in cMYC gene. These data suggest that aberrant expression of A3B can evoke genomic instability by inducing base substitutions into human genome, which might lead to tumorigenesis in human cells.
Related collections
Most cited references39
- Record: found
- Abstract: found
- Article: not found
Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme.
- Record: found
- Abstract: found
- Article: not found