- Record: found
- Abstract: found
- Article: found
Incorporating evolutionary insights to improve ecotoxicology for freshwater species
Read this article at
Abstract
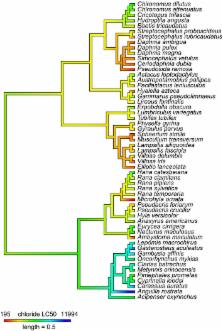
Ecotoxicological studies have provided extensive insights into the lethal and sublethal effects of environmental contaminants. These insights are critical for environmental regulatory frameworks, which rely on knowledge of toxicity for developing policies to manage contaminants. While varied approaches have been applied to ecotoxicological questions, perspectives related to the evolutionary history of focal species or populations have received little consideration. Here, we evaluate chloride toxicity from the perspectives of both macroevolution and contemporary evolution. First, by mapping chloride toxicity values derived from the literature onto a phylogeny of macroinvertebrates, fish, and amphibians, we tested whether macroevolutionary relationships across species and taxa are predictive of chloride tolerance. Next, we conducted chloride exposure tests for two amphibian species to assess whether potential contemporary evolutionary change associated with environmental chloride contamination influences chloride tolerance across local populations. We show that explicitly evaluating both macroevolution and contemporary evolution can provide important and even qualitatively different insights from those obtained via traditional ecotoxicological studies. While macroevolutionary perspectives can help forecast toxicological end points for species with untested sensitivities, contemporary evolutionary perspectives demonstrate the need to consider the environmental context of exposed populations when measuring toxicity. Accounting for divergence among populations of interest can provide more accurate and relevant information related to the sensitivity of populations that may be evolving in response to selection from contaminant exposure. Our data show that approaches accounting for and specifically examining variation among natural populations should become standard practice in ecotoxicology.
Related collections
Most cited references65
- Record: found
- Abstract: found
- Article: not found
Testing for phylogenetic signal in comparative data: behavioral traits are more labile.
- Record: found
- Abstract: not found
- Article: not found
Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments
- Record: found
- Abstract: found
- Article: not found