- Record: found
- Abstract: found
- Article: found
Development of a real-time multiplex PCR assay for the detection of multiple Salmonella serotypes in chicken samples
Read this article at
Abstract
Background
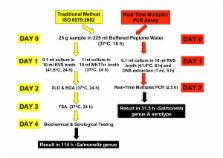
A real-time multiplex PCR assay was developed for the detection of multiple Salmonella serotypes in chicken samples. Poultry-associated serotypes detected in the assay include Enteritidis, Gallinarum, Typhimurium, Kentucky and Dublin. The traditional cultural method according to EN ISO 6579:2002 for the detection of Salmonella in food was performed in parallel. The real-time PCR based method comprised a pre-enrichment step in Buffered Peptone Water (BPW) overnight, followed by a shortened selective enrichment in Rappaport Vasilliadis Soya Broth (RVS) for 6 hours and subsequent DNA extraction.
Results
The real-time multiplex PCR assay and traditional cultural method showed 100% inclusivity and 100% exclusivity on all strains tested. The real-time multiplex PCR assay was as sensitive as the traditional cultural method in detecting Salmonella in artificially contaminated chicken samples and correctly identified the serotype. Artificially contaminated chicken samples resulted in a detection limit of between 1 and 10 CFU per 25 g sample for both methods. A total of sixty-three naturally contaminated chicken samples were investigated by both methods and relative accuracy, relative sensitivity and relative specificity of the real-time PCR method were determined to be 89, 94 and 87%, respectively. Thirty cultures blind tested were correctly identified by the real-time multiplex PCR method.
Related collections
Most cited references37
- Record: found
- Abstract: not found
- Article: not found
Inhibition and facilitation of nucleic acid amplification.
- Record: found
- Abstract: found
- Article: not found
Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions.
- Record: found
- Abstract: found
- Article: not found