- Record: found
- Abstract: found
- Article: found
Isolation and characterization of a new cold-active protease from psychrotrophic bacteria of Western Himalayan glacial soil
Read this article at
Abstract
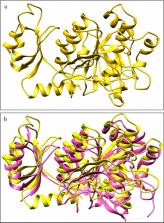
As an approach to the exploration of cold-active enzymes, in this study, we isolated a cold-active protease produced by psychrotrophic bacteria from glacial soils of Thajwas Glacier, Himalayas. The isolated strain BO1, identified as Bacillus pumilus, grew well within a temperature range of 4–30 °C. After its qualitative and quantitative screening, the cold-active protease (Apr-BO1) was purified. The Apr-BO1 had a molecular mass of 38 kDa and showed maximum (37.02 U/mg) specific activity at 20 °C, with casein as substrate. It was stable and active between the temperature range of 5–35 °C and pH 6.0–12.0, with an optimum temperature of 20 °C at pH 9.0. The Apr-BO1 had low K m value of 1.0 mg/ml and V max 10.0 µmol/ml/min. Moreover, it displayed better tolerance to organic solvents, surfactants, metal ions and reducing agents than most alkaline proteases. The results exhibited that it effectively removed the stains even in a cold wash and could be considered a decent detergent additive. Furthermore, through protein modelling, the structure of this protease was generated from template, subtilisin E of Bacillus subtilis (PDB ID: 3WHI), and different methods checked its quality. For the first time, this study reported the protein sequence for psychrotrophic Apr-BO1 and brought forth its novelty among other cold-active proteases.
Related collections
Most cited references87
- Record: found
- Abstract: found
- Article: not found
UCSF Chimera--a visualization system for exploratory research and analysis.
- Record: found
- Abstract: found
- Article: not found
MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets.
- Record: found
- Abstract: not found
- Article: not found