- Record: found
- Abstract: found
- Article: found
Microbial culturomics unravels the halophilic microbiota repertoire of table salt: description of Gracilibacillus massiliensis sp. nov.
Read this article at
Abstract
Background
Microbial culturomics represents an ongoing revolution in the characterization of environmental and human microbiome.
Methods
By using three media containing high salt concentration (100, 150, and 200 g/L), the halophilic microbial culturome of a commercial table salt was determined.
Results
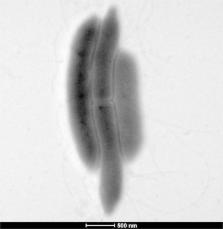
Eighteen species belonging to the Terrabacteria group were isolated including eight moderate halophilic and 10 halotolerant bacteria. Gracilibacillus massiliensis sp. nov., type strain Awa-1 T (=CSUR P1441=DSM 29726), is a moderately halophilic gram-positive, non-spore-forming rod, and is motile by using a flagellum. Strain Awa-1 T shows catalase activity but no oxidase activity. It is not only an aerobic bacterium but also able to grow in anaerobic and microaerophilic atmospheres. The draft genome of G. massiliensis is 4,207,226 bp long, composed of 13 scaffolds with 36.05% of G+C content. It contains 3,908 genes (3,839 protein-coding and 69 RNA genes). At least 1,983 (52%) orthologous proteins were not shared with the closest phylogenetic species. Hundred twenty-six genes (3.3%) were identified as ORFans.
Conclusions
Microbial culturomics can dramatically improve the characterization of the food and environmental microbiota repertoire, deciphering new bacterial species and new genes. Further studies will clarify the geographic specificity and the putative role of these new microbes and their related functional genetic content in environment, health, and disease.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: not found
Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya.
- Record: found
- Abstract: found
- Article: not found