- Record: found
- Abstract: found
- Article: found
Salp blooms drive strong increases in passive carbon export in the Southern Ocean
Read this article at
Abstract
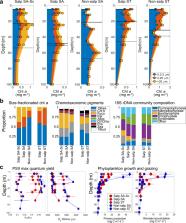
The Southern Ocean contributes substantially to the global biological carbon pump (BCP). Salps in the Southern Ocean, in particular Salpa thompsoni, are important grazers that produce large, fast-sinking fecal pellets. Here, we quantify the salp bloom impacts on microbial dynamics and the BCP, by contrasting locations differing in salp bloom presence/absence. Salp blooms coincide with phytoplankton dominated by diatoms or prymnesiophytes, depending on water mass characteristics. Their grazing is comparable to microzooplankton during their early bloom, resulting in a decrease of ~1/3 of primary production, and negative phytoplankton rates of change are associated with all salp locations. Particle export in salp waters is always higher, ranging 2- to 8- fold (average 5-fold), compared to non-salp locations, exporting up to 46% of primary production out of the euphotic zone. BCP efficiency increases from 5 to 28% in salp areas, which is among the highest recorded in the global ocean.
Abstract
Gelatinous bloom-forming zooplankton—salps—alter microbial communities and quintuple the flux of sinking particles from the surface to the deep, strongly enhancing the ability of the ocean to sequester CO 2.
Related collections
Most cited references85

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
DADA2: High resolution sample inference from Illumina amplicon data

- Record: found
- Abstract: found
- Article: found