- Record: found
- Abstract: found
- Article: found
Feeding an unsalable carrot total-mixed ration altered bacterial amino acid degradation in the rumen of lambs
Read this article at
Abstract
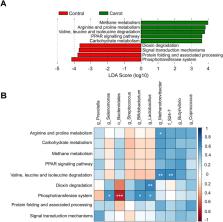
The objective of this study was to determine the influence of a total-mixed ration including unsalable carrots at 45% DM on the rumen microbiome; and the plasma, rumen and liver metabolomes. Carrots discarded at processing were investigated as an energy-dense substitute for barley grain in a conventional feedlot diet, and improved feed conversion efficiency by 25%. Here, rumen fluid was collected from 34 Merino lambs at slaughter (n = 16 control; n = 18 carrot) after a feeding period of 11-weeks. The V4 region of the 16S rRNA gene was sequenced to profile archaeal and bacterial microbe communities. Further, a comprehensive, targeted profile of known metabolites was constructed for blood plasma, rumen fluid and biopsied liver metabolites using a gas chromatography mass spectrometry (GC–MS) metabolomics approach. An in vitro batch culture was used to characterise ruminal fermentation including gas and methane (CH 4) production. In vivo rumen microbial community structure of carrot fed lambs was dissimilar ( P < 0.01; PERMANOVA), and all measures of alpha diversity were greater ( P < 0.01), compared to those fed the control diet. Unclassified genera in Bacteroidales (15.9 ± 6.74% relative abundance; RA) were more abundant ( P < 0.01) in the rumen fluid of carrot-fed lambs, while unclassified taxa in the Succinivibrionaceae family (11.1 ± 3.85% RA) were greater ( P < 0.01) in the control. The carrot diet improved in vitro ruminal fermentation evidenced as an 8% increase ( P < 0.01) in DM digestibility and a 13.8% reduction ( P = 0.01) in CH 4 on a mg/ g DM basis, while the control diet increased ( P = 0.04) percentage of propionate within total VFA by 20%. Fourteen rumen fluid metabolites and 27 liver metabolites were influenced ( P ≤ 0.05) by diet, while no effect ( P ≥ 0.05) was observed in plasma metabolites. The carrot diet enriched (impact value = 0.13; P = 0.01) the tyrosine metabolism pathway (acetoacetic acid, dopamine and pyruvate), while the control diet enriched (impact value = 0.42; P ≤ 0.02) starch and sucrose metabolism (trehalose and glucose) in rumen fluid. This study demonstrated that feeding 45% DM unsalable carrots diversified bacterial communities in the rumen. These dietary changes influenced pathways of tyrosine degradation, such that previous improvements in feed conversion efficiency in lambs could be explained.
Related collections
Most cited references96
- Record: found
- Abstract: found
- Article: not found
DADA2: High resolution sample inference from Illumina amplicon data
- Record: found
- Abstract: found
- Article: not found
KEGG: kyoto encyclopedia of genes and genomes.

- Record: found
- Abstract: found
- Article: found