- Record: found
- Abstract: found
- Article: found
Promoter activity dynamics in the lag phase of Escherichia coli
Read this article at
Abstract
Background
Lag phase is a period of time with no growth that occurs when stationary phase bacteria are transferred to a fresh medium. Bacteria in lag phase seem inert: their biomass does not increase. The low number of cells and low metabolic activity make it difficult to study this phase. As a consequence, it has not been studied as thoroughly as other bacterial growth phases. However, lag phase has important implications for bacterial infections and food safety. We asked which, if any, genes are expressed in the lag phase of Escherichia coli, and what is their dynamic expression pattern.
Results
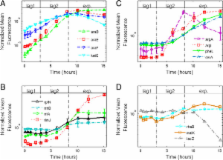
We developed an assay based on imaging flow cytometry of fluorescent reporter cells that overcomes the challenges inherent in studying lag phase. We distinguish between lag1 phase- in which there is no biomass growth, and lag2 phase- in which there is biomass growth but no cell division. We find that in lag1 phase, most promoters are not active, except for the enzymes that utilize the specific carbon source in the medium. These genes show promoter activities that increase exponentially with time, despite the fact that the cells do not measurably increase in size. An oxidative stress promoter, katG, is also active. When cells enter lag2 and begin to grow in size, they switch to a full growth program of promoter activity including ribosomal and metabolic genes.
Conclusions
The observed exponential increase in enzymes for the specific carbon source followed by an abrupt switch to production of general growth genes is a solution of an optimal control model, known as bang-bang control. The present approach contributes to the understanding of lag phase, the least studied of bacterial growth phases.
Related collections
Most cited references60
- Record: found
- Abstract: found
- Article: not found
Stochasticity in gene expression: from theories to phenotypes.
- Record: found
- Abstract: found
- Article: not found
A dynamic approach to predicting bacterial growth in food.
- Record: found
- Abstract: found
- Article: not found