- Record: found
- Abstract: found
- Article: found
DNA origami scaffold for studying intrinsically disordered proteins of the nuclear pore complex
Read this article at
Abstract
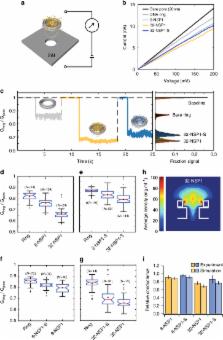
The nuclear pore complex (NPC) is the gatekeeper for nuclear transport in eukaryotic cells. A key component of the NPC is the central shaft lined with intrinsically disordered proteins (IDPs) known as FG-Nups, which control the selective molecular traffic. Here, we present an approach to realize artificial NPC mimics that allows controlling the type and copy number of FG-Nups. We constructed 34 nm-wide 3D DNA origami rings and attached different numbers of NSP1, a model yeast FG-Nup, or NSP1-S, a hydrophilic mutant. Using (cryo) electron microscopy, we find that NSP1 forms denser cohesive networks inside the ring compared to NSP1-S. Consistent with this, the measured ionic conductance is lower for NSP1 than for NSP1-S. Molecular dynamics simulations reveal spatially varying protein densities and conductances in good agreement with the experiments. Our technique provides an experimental platform for deciphering the collective behavior of IDPs with full control of their type and position.
Abstract
FG-Nups are disordered proteins in the nuclear pore complex (NPC) where they selectively control nuclear transport. Here authors build NPC-mimics based on DNA origami rings which attach a certain numbers of Nups to analyse those nanopores by cryoEM and conductance measurements.
Related collections
Most cited references37
- Record: found
- Abstract: found
- Article: not found
Folding DNA into twisted and curved nanoscale shapes.
