- Record: found
- Abstract: found
- Article: found
BioJava: an open-source framework for bioinformatics
Read this article at
Abstract
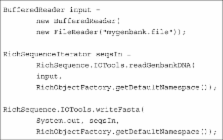
Summary: BioJava is a mature open-source project that provides a framework for processing of biological data. BioJava contains powerful analysis and statistical routines, tools for parsing common file formats and packages for manipulating sequences and 3D structures. It enables rapid bioinformatics application development in the Java programming language.
Availability: BioJava is an open-source project distributed under the Lesser GPL (LGPL). BioJava can be downloaded from the BioJava website ( http://www.biojava.org). BioJava requires Java 1.5 or higher.
Contact: andreas.prlic@ 123456gmail.com . All queries should be directed to the BioJava mailing lists. Details are available at http://biojava.org/wiki/BioJava:MailingLists.
Related collections
Most cited references5
- Record: found
- Abstract: found
- Article: not found
The Bioperl toolkit: Perl modules for the life sciences.
- Record: found
- Abstract: found
- Article: not found
The Bio* toolkits--a brief overview.
- Record: found
- Abstract: not found
- Article: not found