- Record: found
- Abstract: found
- Article: found
Genetic variation and evolutionary history of a mycorrhizal fungus regulate the currency of exchange in symbiosis with the food security crop cassava
Read this article at
Abstract
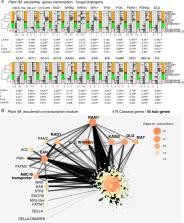
Most land plants form symbioses with arbuscular mycorrhizal fungi (AMF). Diversity of AMF increases plant community productivity and plant diversity. For decades, it was known that plants trade carbohydrates for phosphate with their fungal symbionts. However, recent studies show that plant-derived lipids probably represent the most essential currency of exchange. Understanding the regulation of plant genes involved in the currency of exchange is crucial to understanding stability of this mutualism. Plants encounter many different AMF genotypes that vary greatly in the benefit they confer to plants. Yet the role that fungal genetic variation plays in the regulation of this currency has not received much attention. We used a high-resolution phylogeny of one AMF species ( Rhizophagus irregularis) to show that fungal genetic variation drives the regulation of the plant fatty acid pathway in cassava ( Manihot esculenta); a pathway regulating one of the essential currencies of trade in the symbiosis. The regulation of this pathway was explained by clearly defined patterns of fungal genome-wide variation representing the precise fungal evolutionary history. This represents the first demonstrated link between the genetics of AMF and reprogramming of an essential plant pathway regulating the currency of exchange in the symbiosis. The transcription factor RAM1 was also revealed as the dominant gene in the fatty acid plant gene co-expression network. Our study highlights the crucial role of variation in fungal genomes in the trade of resources in this important symbiosis and also opens the door to discovering characteristics of AMF genomes responsible for interactions between AMF and cassava that will lead to optimal cassava growth.
Related collections
Most cited references34
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: found
- Article: not found
Testing for phylogenetic signal in comparative data: behavioral traits are more labile.
- Record: found
- Abstract: found
- Article: not found