- Record: found
- Abstract: found
- Article: found
Discovering Digital Tumor Signatures—Using Latent Code Representations to Manipulate and Classify Liver Lesions
Read this article at
Abstract
Simple Summary
We use a generative deep learning paradigm for the identification of digital signatures in radiological imaging data. The model is trained on a small inhouse data set and evaluated on publicly available data. Apart from using the learned signatures for the characterization of lesions, in analogy to radiomics features, we also demonstrate that by manipulating them we can create realistic synthetic CT image patches. This generation of synthetic data can be carried out at user-defined spatial locations. Moreover, the discrimination of liver lesions from normal liver tissue can be achieved with high accuracy, sensitivity, and specificity.
Abstract
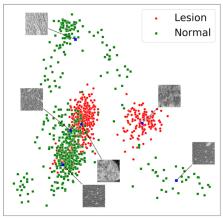
Modern generative deep learning (DL) architectures allow for unsupervised learning of latent representations that can be exploited in several downstream tasks. Within the field of oncological medical imaging, we term these latent representations “digital tumor signatures” and hypothesize that they can be used, in analogy to radiomics features, to differentiate between lesions and normal liver tissue. Moreover, we conjecture that they can be used for the generation of synthetic data, specifically for the artificial insertion and removal of liver tumor lesions at user-defined spatial locations in CT images. Our approach utilizes an implicit autoencoder, an unsupervised model architecture that combines an autoencoder and two generative adversarial network (GAN)-like components. The model was trained on liver patches from 25 or 57 inhouse abdominal CT scans, depending on the experiment, demonstrating that only minimal data is required for synthetic image generation. The model was evaluated on a publicly available data set of 131 scans. We show that a PCA embedding of the latent representation captures the structure of the data, providing the foundation for the targeted insertion and removal of tumor lesions. To assess the quality of the synthetic images, we conducted two experiments with five radiologists. For experiment 1, only one rater and the ensemble-rater were marginally above the chance level in distinguishing real from synthetic data. For the second experiment, no rater was above the chance level. To illustrate that the “digital signatures” can also be used to differentiate lesion from normal tissue, we employed several machine learning methods. The best performing method, a LinearSVM, obtained 95% (97%) accuracy, 94% (95%) sensitivity, and 97% (99%) specificity, depending on if all data or only normal appearing patches were used for training of the implicit autoencoder. Overall, we demonstrate that the proposed unsupervised learning paradigm can be utilized for the removal and insertion of liver lesions at user defined spatial locations and that the digital signatures can be used to discriminate between lesions and normal liver tissue in abdominal CT scans.
Related collections
Most cited references36
- Record: found
- Abstract: not found
- Article: not found
Extracting and composing robust features with denoising autoencoders
- Record: found
- Abstract: not found
- Article: not found
GAN-based Synthetic Medical Image Augmentation for increased CNN Performance in Liver Lesion Classification

- Record: found
- Abstract: found
- Article: found