- Record: found
- Abstract: found
- Article: found
ER-phagy: mechanisms, regulation, and diseases connected to the lysosomal clearance of the endoplasmic reticulum
Read this article at
Abstract
Abstract
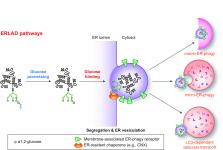
ER-phagy (reticulophagy) defines the degradation of portions of the endoplasmic reticulum (ER) within lysosomes or vacuoles. It is part of the self-digestion (i.e., autophagic) programs recycling cytoplasmic material and organelles, which rapidly mobilize metabolites in cells confronted with nutrient shortage. Moreover, selective clearance of ER subdomains participates in the control of ER size and activity during ER stress, the reestablishment of ER homeostasis after ER stress resolution, and the removal of ER parts in which aberrant and potentially cytotoxic material has been segregated. ER-phagy relies on the individual and/or concerted activation of the ER-phagy receptors, ER peripheral or integral membrane proteins that share the presence of LC3/Atg8-binding motifs in their cytosolic domains. ER-phagy involves the physical separation of portions of the ER from the bulk ER network and their delivery to the endolysosomal/vacuolar catabolic district. This last step is accomplished by a variety of mechanisms including macro-ER-phagy (in which ER fragments are sequestered by double-membrane autophagosomes that eventually fuse with lysosomes/vacuoles), micro-ER-phagy (in which ER fragments are directly engulfed by endosomes/lysosomes/vacuoles), or direct fusion of ER-derived vesicles with lysosomes/vacuoles. ER-phagy is dysfunctional in specific human diseases, and its regulators are subverted by pathogens, highlighting its crucial role for cell and organism life.
Related collections
Most cited references545
- Record: found
- Abstract: found
- Article: not found
Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans.
- Record: found
- Abstract: not found
- Article: not found
Mechanism and medical implications of mammalian autophagy
- Record: found
- Abstract: found
- Article: not found
