- Record: found
- Abstract: found
- Article: found
Tracing the history of LINE and SINE extinction in sigmodontine rodents
Read this article at
Abstract
Background
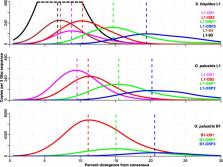
L1 retrotransposons have co-evolved with their mammalian hosts for the entire history of mammals and currently compose ~ 20% of a mammalian genome. B1 retrotransposons are dependent on L1 for retrotransposition and span the evolutionary history of rodents since their radiation. L1s were found to have lost their activity in a group of South American rodents, the Sigmodontinae, and B1 inactivation preceded the extinction of L1 in the same group. Consequently, a basal group of sigmodontines have active L1s but inactive B1s and a derived clade have both inactive L1s and B1s. It has been suggested that B1s became extinct during a long period of L1 quiescence and that L1s subsequently reemerged in the basal group.
Results
Here we investigate the evolutionary histories of L1 and B1 in the sigmodontine rodents and show that L1 activity continued until after the L1-extinct clade and the basal group diverged. After the split, L1 had a small burst of activity in the former group, followed by extinction. In the basal group, activity was initially low but was followed by a dramatic increase in L1 activity. We found the last wave of B1 retrotransposition was large and probably preceded the split between the two rodent clades.
Conclusions
Given that L1s had been steadily retrotransposing during the time corresponding to B1 extinction and that the burst of B1 activity preceding B1 extinction was large, we conclude that B1 extinction was not a result of L1 quiescence. Rather, the burst of B1 activity may have contributed to L1 extinction both by competition with L1 and by putting strong selective pressure on the host to control retrotransposition.
Related collections
Most cited references53
- Record: found
- Abstract: found
- Article: not found
FLASH: fast length adjustment of short reads to improve genome assemblies.
- Record: found
- Abstract: found
- Article: not found
L1 retrotransposition in human neural progenitor cells.
- Record: found
- Abstract: found
- Article: not found