- Record: found
- Abstract: found
- Article: found
Grass Carp Laboratory of Genetics and Physiology 2 Serves As a Negative Regulator in Retinoic Acid-Inducible Gene I- and Melanoma Differentiation-Associated Gene 5-Mediated Antiviral Signaling in Resting State and Early Stage of Grass Carp Reovirus Infection
Read this article at
Abstract
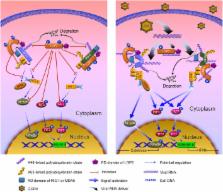
Laboratory of genetics and physiology 2 (LGP2) is a key component of RIG-I-like receptors (RLRs). However, the lack of the caspase recruitment domains (CARDs) results in its controversial functional performance as a negative or positive regulator in antiviral responses. Especially, no sufficient evidence uncovers the functional mechanisms of LGP2 in RLR signaling pathways in teleost. Here, negative regulation mechanism of LGP2 in certain situations in retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5)-mediated antiviral responses was identified in Ctenopharyngodon idella kidney cells. LGP2 overexpression inhibits synthesis and phosphorylation of interferon regulatory factor 3/7 (IRF3/7), and mRNA levels and promoter activities of IFNs and NF-κBs in resting state and early phase of grass carp reovirus (GCRV) infection. Knockdown of LGP2 obtains opposite effects. Luciferase report assay indicates that LGP2 works at the upstream of RIG-I and MDA5. LGP2 binds to RIG-I and MDA5 with diverse domain preference and which is independent of GCRV infection. Furthermore, LGP2 restrains K63-linked ubiquitination of RIG-I and MDA5 in various degrees. These differences result in disparate repressive mechanisms of LGP2 to RIG-I- and MDA5-mediated signal activations of IFN-β promoter stimulator 1 and mediator of IRF3 activation. Interestingly, LGP2 also inhibits K48-linked RIG-I and MDA5 ubiquitination to suppress proteins degradation, which guarantees the basal protein levels for subsequently rapid signal activation. All these results reveal a mechanism that LGP2 functions as a suppressor in RLR signaling pathways to maintain cellular homeostasis in resting state and early phase during GCRV infection.
Related collections
Most cited references50
- Record: found
- Abstract: found
- Article: not found
Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA.
- Record: found
- Abstract: found
- Article: not found
Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2.
- Record: found
- Abstract: found
- Article: not found