- Record: found
- Abstract: found
- Article: found
The genome sequence of Propionibacterium acidipropionici provides insights into its biotechnological and industrial potential
Read this article at
Abstract
Background
Synthetic biology allows the development of new biochemical pathways for the production of chemicals from renewable sources. One major challenge is the identification of suitable microorganisms to hold these pathways with sufficient robustness and high yield. In this work we analyzed the genome of the propionic acid producer Actinobacteria Propionibacterium acidipropionici (ATCC 4875).
Results
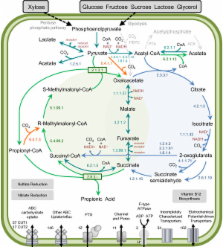
The assembled P. acidipropionici genome has 3,656,170 base pairs (bp) with 68.8% G + C content and a low-copy plasmid of 6,868 bp. We identified 3,336 protein coding genes, approximately 1000 more than P. freudenreichii and P. acnes, with an increase in the number of genes putatively involved in maintenance of genome integrity, as well as the presence of an invertase and genes putatively involved in carbon catabolite repression. In addition, we made an experimental confirmation of the ability of P. acidipropionici to fix CO 2, but no phosphoenolpyruvate carboxylase coding gene was found in the genome. Instead, we identified the pyruvate carboxylase gene and confirmed the presence of the corresponding enzyme in proteome analysis as a potential candidate for this activity. Similarly, the phosphate acetyltransferase and acetate kinase genes, which are considered responsible for acetate formation, were not present in the genome. In P. acidipropionici, a similar function seems to be performed by an ADP forming acetate-CoA ligase gene and its corresponding enzyme was confirmed in the proteome analysis.
Conclusions
Our data shows that P. acidipropionici has several of the desired features that are required to become a platform for the production of chemical commodities: multiple pathways for efficient feedstock utilization, ability to fix CO 2, robustness, and efficient production of propionic acid, a potential precursor for valuable 3-carbon compounds.
Related collections
Most cited references63
- Record: found
- Abstract: found
- Article: not found
UniRef: comprehensive and non-redundant UniProt reference clusters.
- Record: found
- Abstract: found
- Article: not found