- Record: found
- Abstract: found
- Article: found
Indole-3-lactic acid associated with Bifidobacterium-dominated microbiota significantly decreases inflammation in intestinal epithelial cells
Read this article at
Abstract
Background
Bifidobacterium longum subsp. infantis ( B. infantis) is a commensal bacterium that colonizes the gastrointestinal tract of breast-fed infants. B. infantis can efficiently utilize the abundant supply of oligosaccharides found in human milk (HMO) to help establish residence. We hypothesized that metabolites from B. infantis grown on HMO produce a beneficial effect on the host.
Results
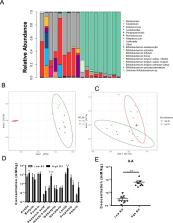
In a previous study, we demonstrated that B. infantis routinely dominated the fecal microbiota of a breast fed Bangladeshi infant cohort (1). Characterization of the fecal metabolome of binned samples representing high and low B. infantis populations from this cohort revealed higher amounts of the tryptophan metabolite indole-3-lactic acid (ILA) in feces with high levels of B. infantis. Further in vitro analysis confirmed that B. infantis produced significantly greater quantities of the ILA when grown on HMO versus lactose, suggesting a growth substrate relationship to ILA production. The direct effects of ILA were assessed in a macrophage cell line and intestinal epithelial cell lines. ILA (1-10 mM) significantly attenuated lipopolysaccharide (LPS)-induced activation of NF-kB in macrophages. ILA significantly attenuated TNF-α- and LPS-induced increase in the pro-inflammatory cytokine IL-8 in intestinal epithelial cells. ILA increased mRNA expression of the aryl hydrogen receptor (AhR)-target gene CYP1A1 and nuclear factor erythroid 2–related factor 2 (Nrf2)-targeted genes glutathione reductase 2 (GPX2), superoxide dismutase 2 (SOD2), and NAD(P) H dehydrogenase (NQO1). Pretreatment with either the AhR antagonist or Nrf-2 antagonist inhibited the response of ILA on downstream effectors.
Related collections
Most cited references72
- Record: found
- Abstract: not found
- Article: not found
QIIME allows analysis of high-throughput community sequencing data.

- Record: found
- Abstract: found
- Article: found
phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data

- Record: found
- Abstract: found
- Article: found
