- Record: found
- Abstract: found
- Article: found
Release of transcriptional repression through the HCR promoter region confers uniform expression of HWP1 on surfaces of Candida albicans germ tubes
Read this article at
Abstract
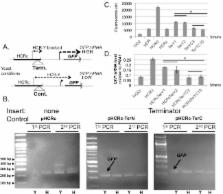
The mechanisms that fungi use to co-regulate subsets of genes specifically associated with morphogenic states represent a basic unsolved problem in fungal biology. Candida albicans is an important model of fungal differentiation both for rapid interconversion between yeast and hyphal growth forms and for white/opaque switching mechanisms. The Sundstrom lab is interested in mechanisms regulating hypha-specific expression of adhesin genes that are critical for C. albicans hyphal growth phenotypes and pathogenicity. Early studies on hypha-specific genes such as HWP1 and ALS3 reported 5’ intergenic regions that are larger than those typically found in an average promoter and are associated with hypha-specific expression. In the case of HWP1, activation and repression involves a 368 bp region, denoted the HWP1 control region (HCR), located 1410 bp upstream of its transcription start site. In previous work we showed that HCR confers developmental regulation to a heterologous ENO1 promoter, indicating that HCR by itself contains sufficient information to couple gene expression to morphology. Here we show that the activation and repression mediated by HCR are localized to distinct HCR regions that are targeted by the transcription factors Nrg1p and Efg1p. The finding that Efg1p mediates both repression via HCR under yeast morphological conditions and activation conditions positions Efg1p as playing a central role in coupling HWP1 expression to morphogenesis through the HCR region. These localization studies revealed that the 120 terminal base pairs of HCR confer Efg1p-dependent repressive activity in addition to the Nrg1p repressive activity mediated by DNA upstream of this subregion. The 120 terminal base pair subregion of HCR also contained an initiation site for an HWP1 transcript that is specific to yeast growth conditions (HCR-Y) and may function in the repression of downstream DNA. The detection of an HWP1 mRNA isoform specific to hyphal growth conditions (HWP1-H) showed that morphology-specific mRNA isoforms occur under both yeast and hyphal growth conditions. Similar results were found at the ALS3 locus. Taken together, these results, suggest that the long 5’ intergenic regions upstream of hypha-specific genes function in generating mRNA isoforms that are important for morphology-specific gene expression. Additional complexity in the HWP1 promoter involving HCR-independent activation was discovered by creating a strain lacking HCR that exhibited variable HWP1 expression during hyphal growth conditions. These results show that while HCR is important for ensuring uniform HWP1 expression in cell populations, HCR independent expression also exists. Overall, these results elucidate HCR-dependent mechanisms for coupling HWP1-dependent gene expression to morphology uniformly in cell populations and prompt the hypothesis that mRNA isoforms may play a role in coupling gene expression to morphology in C. albicans.
Related collections
Most cited references52
- Record: found
- Abstract: found
- Article: not found
Nonfilamentous C. albicans mutants are avirulent.
- Record: found
- Abstract: found
- Article: not found
Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1.
- Record: found
- Abstract: found
- Article: not found