- Record: found
- Abstract: found
- Article: found
Plant Organelle Genome Replication
Read this article at
Abstract
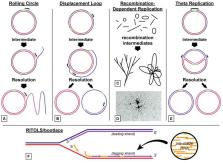
Mitochondria and chloroplasts perform essential functions in respiration, ATP production, and photosynthesis, and both organelles contain genomes that encode only some of the proteins that are required for these functions. The proteins and mechanisms for organelle DNA replication are very similar to bacterial or phage systems. The minimal replisome may consist of DNA polymerase, a primase/helicase, and a single-stranded DNA binding protein (SSB), similar to that found in bacteriophage T7. In Arabidopsis, there are two genes for organellar DNA polymerases and multiple potential genes for SSB, but there is only one known primase/helicase protein to date. Genome copy number varies widely between type and age of plant tissues. Replication mechanisms are only poorly understood at present, and may involve multiple processes, including recombination-dependent replication (RDR) in plant mitochondria and perhaps also in chloroplasts. There are still important questions remaining as to how the genomes are maintained in new organelles, and how genome copy number is determined. This review summarizes our current understanding of these processes.
Related collections
Most cited references117
- Record: found
- Abstract: not found
- Article: not found
Comparative organization of chloroplast genomes.
- Record: found
- Abstract: found
- Article: not found
Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates.
- Record: found
- Abstract: not found
- Article: not found