- Record: found
- Abstract: found
- Article: found
Human Cytomegalovirus Tegument Protein pUL71 Is Required for Efficient Virion Egress
Read this article at
Abstract
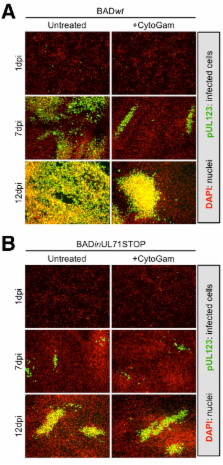
The human cytomegalovirus virion is composed of a DNA genome packaged in an icosahedral capsid, surrounded by a tegument of protein and RNA, all enclosed within a glycoprotein-studded envelope. Achieving this intricate virion architecture requires a coordinated process of assembly and egress. We show here that pUL71, a component of the virion tegument with a previously uncharacterized function, is required for the virus-induced reorganization of host cell membranes, which is necessary for efficient viral assembly and egress. A mutant that did not express pUL71 was able to efficiently accumulate viral genomes and proteins that were tested but was defective for the production and release of infectious virions. The protein localized to vesicular structures at the periphery of the viral assembly compartment, and during infection with a pUL71-deficient virus, these structures were grossly enlarged and aberrantly contained a cellular marker of late endosomes/lysosomes. Mutant virus preparations exhibited less infectivity per unit genome than wild-type virus preparations, due to aggregation of virus particles and their association with membrane fragments. Finally, mutant virus particles accumulated within the cytoplasm of infected cells and were localized to the periphery of large structures with properties of lysosomes, whose formation was kinetically favored in mutant-virus-infected cells. Together, these observations point to a role for pUL71 in the establishment and/or maintenance of a functional viral assembly compartment that is required for normal virion trafficking and egress from infected cells.
IMPORTANCE
In addition to causing disease in immunocompromised individuals, human cytomegalovirus is the leading known infectious cause of birth defects. To induce these pathologies, the virus must spread from its site of introduction to various organs and tissues in the body. The processes of viral assembly and egress, which underlie the spread of infection, are incompletely understood. We elucidate a role for a virus-coded protein, pUL71, in these processes and demonstrate the importance of maintaining an intricate, virus-induced reorganization of host cell membranes for efficient virus spread.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
A highly efficient recombineering-based method for generating conditional knockout mutations.

- Record: found
- Abstract: found
- Article: found
CDD: specific functional annotation with the Conserved Domain Database
- Record: found
- Abstract: found
- Article: not found