- Record: found
- Abstract: found
- Article: found
Neutralising antibody response in domestic cats immunised with a commercial feline immunodeficiency virus (FIV) vaccine
Read this article at
Highlights
Abstract
Across human and veterinary medicine, vaccines against only two retroviral infections have been brought to market successfully, the vaccines against feline leukaemia virus (FeLV) and feline immunodeficiency virus (FIV). FeLV vaccines have been a global success story, reducing virus prevalence in countries where uptake is high. In contrast, the more recent FIV vaccine was introduced in 2002 and the degree of protection afforded in the field remains to be established. However, given the similarities between FIV and HIV, field studies of FIV vaccine efficacy are likely to advise and inform the development of future approaches to HIV vaccination.
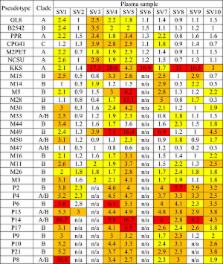
Here we assessed the neutralising antibody response induced by FIV vaccination against a panel of FIV isolates, by testing blood samples collected from client-owned vaccinated Australian cats. We examined the molecular and phenotypic properties of 24 envs isolated from one vaccinated cat that we speculated might have become infected following natural exposure to FIV. Cats vaccinated against FIV did not display broadly neutralising antibodies, suggesting that protection may not extend to some virulent recombinant strains of FIV circulating in Australia.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination.
- Record: found
- Abstract: found
- Article: not found
Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology.
- Record: found
- Abstract: not found
- Article: not found