- Record: found
- Abstract: found
- Article: found
Single-cell molecular profiling of all three components of the HPA axis reveals adrenal ABCB1 as a regulator of stress adaptation
Read this article at
Abstract
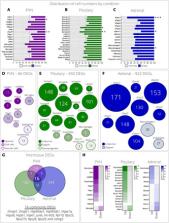
Dissection of the HPA axis using single-cell RNA sequencing uncovers adrenal ABCB1 as an important player of stress adaptation.
Abstract
Chronic activation and dysregulation of the neuroendocrine stress response have severe physiological and psychological consequences, including the development of metabolic and stress-related psychiatric disorders. We provide the first unbiased, cell type–specific, molecular characterization of all three components of the hypothalamic-pituitary-adrenal axis, under baseline and chronic stress conditions. Among others, we identified a previously unreported subpopulation of Abcb1b + cells involved in stress adaptation in the adrenal gland. We validated our findings in a mouse stress model, adrenal tissues from patients with Cushing’s syndrome, adrenocortical cell lines, and peripheral cortisol and genotyping data from depressed patients. This extensive dataset provides a valuable resource for researchers and clinicians interested in the organism’s nervous and endocrine responses to stress and the interplay between these tissues. Our findings raise the possibility that modulating ABCB1 function may be important in the development of treatment strategies for patients suffering from metabolic and stress-related psychiatric disorders.
Related collections
Most cited references68
- Record: found
- Abstract: found
- Article: found
SCANPY : large-scale single-cell gene expression data analysis
- Record: found
- Abstract: found
- Article: not found