- Record: found
- Abstract: found
- Article: found
Regulation of ENaC-Mediated Sodium Reabsorption by Peroxisome Proliferator-Activated Receptors
Read this article at
Abstract
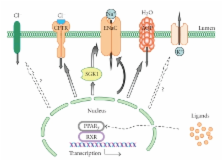
Peroxisome proliferator-activated receptors (PPARs) are members of a steroid hormone receptor superfamily that responds to changes in lipid and glucose homeostasis. Peroxisomal proliferator-activated receptor subtype γ (PPAR γ) has received much attention as the target for antidiabetic drugs, as well as its role in responding to endogenous compounds such as prostaglandin J 2. However, thiazolidinediones (TZDs), the synthetic agonists of the PPAR γ are tightly associated with fluid retention and edema, as potentially serious side effects. The epithelial sodium channel (ENaC) represents the rate limiting step for sodium absorption in the renal collecting duct. Consequently, ENaC is a central effector impacting systemic blood volume and pressure. The role of PPAR γ agonists on ENaC activity remains controversial. While PPAR γ agonists were shown to stimulate ENaC-mediated renal salt absorption, probably via Serum- and Glucocorticoid-Regulated Kinase 1 (SGK1), other studies reported that PPAR γ agonist-induced fluid retention is independent of ENaC activity. The current paper provides new insights into the control and function of ENaC and ENaC-mediated sodium transport as well as several other epithelial channels/transporters by PPARs and particularly PPAR γ. The potential contribution of arachidonic acid (AA) metabolites in PPAR-dependent mechanisms is also discussed.
Related collections
Most cited references89
- Record: found
- Abstract: found
- Article: not found
Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat.
- Record: found
- Abstract: found
- Article: not found