- Record: found
- Abstract: found
- Article: found
Knockdown of MLO genes reduces susceptibility to powdery mildew in grapevine
Read this article at
Abstract
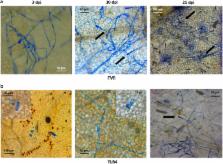
Erysiphe necator is the causal agent of powdery mildew (PM), one of the most destructive diseases of grapevine. PM is controlled by sulfur-based and synthetic fungicides, which every year are dispersed into the environment. This is why PM-resistant varieties should become a priority for sustainable grapevine and wine production. PM resistance can be achieved in other crops by knocking out susceptibility S-genes, such as those residing at genetic loci known as MLO ( Mildew Locus O). All MLO S-genes of dicots belong to the phylogenetic clade V, including grapevine genes VvMLO7, 11 and 13, which are upregulated during PM infection, and VvMLO6, which is not upregulated. Before adopting a gene-editing approach to knockout candidate S-genes, the evidence that loss of function of MLO genes can reduce PM susceptibility is necessary. This paper reports the knockdown through RNA interference of VvMLO6, 7, 11 and 13. The knockdown of VvMLO6, 11 and 13 did not decrease PM severity, whereas the knockdown of VvMLO7 in combination with VvMLO6 and VvMLO11 reduced PM severity up to 77%. The knockdown of VvMLO7 and VvMLO6 seemed to be important for PM resistance, whereas a role for VvMLO11 does not seem likely. Cell wall appositions (papillae) were present in both resistant and susceptible lines in response to PM attack. Thirteen genes involved in defense were less upregulated in infected mlo plants, highlighting the early mlo-dependent disruption of PM invasion.
Related collections
Most cited references35

- Record: found
- Abstract: found
- Article: found
An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development
- Record: found
- Abstract: found
- Article: not found
The barley Mlo gene: a novel control element of plant pathogen resistance.
- Record: found
- Abstract: found
- Article: not found