- Record: found
- Abstract: found
- Article: found
Validating the doubly weighted genetic risk score for the prediction of type 2 diabetes in the Lifelines and Estonian Biobank cohorts
Read this article at
Abstract
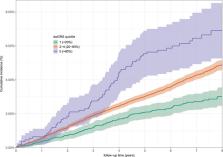
As many cases of type 2 diabetes (T2D) are likely to remain undiagnosed, better tools for early detection of high‐risk individuals are needed to prevent or postpone the disease. We investigated the value of the doubly weighted genetic risk score (dwGRS) for the prediction of incident T2D in the Lifelines and Estonian Biobank (EstBB) cohorts. The dwGRS uses an additional weight for each single nucleotide polymorphism in the risk score, to correct for “Winner's curse” bias in the effect size estimates. The traditional (single‐weighted genetic risk score; swGRS) and dwGRS were calculated for participants in Lifelines ( n = 12,018) and EstBB ( n = 34,129). The dwGRS was found to have stronger association with incident T2D (hazard ratio [HR] = 1.26 [95% confidence interval: 1.10–1.43] and HR = 1.35 [1.28–1.42]) compared to the swGRS (HR = 1.21 [1.07–1.38] and HR = 1.25 [1.19–1.32]) in Lifelines and EstBB, respectively. Comparing the 5‐year predicted risks from the models with and without the dwGRS, the continuous net reclassification index was 0.140 (0.034–0.243; p = .009 Lifelines), and 0.257 (0.194–0.319; p < 2 × 10 −16 EstBB). The dwGRS provided incremental value to the T2D prediction model with established phenotypic predictors. It clearly distinguished the risk groups for incident T2D in both biobanks thereby showing its clinical relevance.
Related collections
Most cited references20
- Record: found
- Abstract: found
- Article: not found
Pathophysiology and treatment of type 2 diabetes: perspectives on the past, present, and future.
- Record: found
- Abstract: found
- Article: not found
Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers.
- Record: found
- Abstract: not found
- Article: not found