- Record: found
- Abstract: found
- Article: found
DeepARV: ensemble deep learning to predict drug-drug interaction of clinical relevance with antiretroviral therapy
Read this article at
Abstract
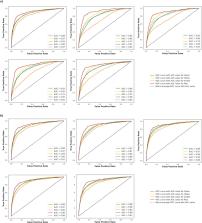
Drug-drug interaction (DDI) may result in clinical toxicity or treatment failure of antiretroviral therapy (ARV) or comedications. Despite the high number of possible drug combinations, only a limited number of clinical DDI studies are conducted. Computational prediction of DDIs could provide key evidence for the rational management of complex therapies. Our study aimed to assess the potential of deep learning approaches to predict DDIs of clinical relevance between ARVs and comedications. DDI severity grading between 30,142 drug pairs was extracted from the Liverpool HIV Drug Interaction database. Two feature construction techniques were employed: 1) drug similarity profiles by comparing Morgan fingerprints, and 2) embeddings from SMILES of each drug via ChemBERTa, a transformer-based model. We developed DeepARV-Sim and DeepARV-ChemBERTa to predict four categories of DDI: i) Red: drugs should not be co-administered, ii) Amber: interaction of potential clinical relevance manageable by monitoring/dose adjustment, iii) Yellow: interaction of weak relevance and iv) Green: no expected interaction. The imbalance in the distribution of DDI severity grades was addressed by undersampling and applying ensemble learning. DeepARV-Sim and DeepARV-ChemBERTa predicted clinically relevant DDI between ARVs and comedications with a weighted mean balanced accuracy of 0.729 ± 0.012 and 0.776 ± 0.011, respectively. DeepARV-Sim and DeepARV-ChemBERTa have the potential to leverage molecular structures associated with DDI risks and reduce DDI class imbalance, effectively increasing the predictive ability on clinically relevant DDIs. This approach could be developed for identifying high-risk pairing of drugs, enhancing the screening process, and targeting DDIs to study in clinical drug development.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
Deep learning.
- Record: found
- Abstract: found
- Article: not found
Extended-connectivity fingerprints.
- Record: found
- Abstract: not found
- Article: not found