- Record: found
- Abstract: found
- Article: not found
Rapid response research to emerging infectious diseases: lessons from SARS
Read this article at
Abstract
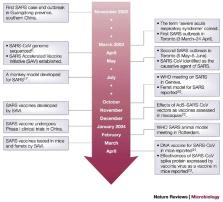
New and emerging infectious diseases continue to plague the world, and there is significant concern that recombinant infectious agents can be used as bioterrorism threats. Microbiologists are increasingly being asked to apply their scientific knowledge to respond to these threats. The recent pandemic caused by the severe acute respiratory syndrome (SARS) coronavirus illustrated not only how a newly evolved pathogen can rapidly spread throughout the world but also how the global community can unite to identify the causative agent and control its spread. Rapid response research mechanisms, such as those used by the SARS Accelerated Vaccine Initiative (SAVI), have shown that the application of emergency management techniques, together with rapid response research, can be highly effective when applied appropriately to new infectious diseases.
Related collections
Most cited references14
- Record: found
- Abstract: found
- Article: not found
Characterization of a novel coronavirus associated with severe acute respiratory syndrome.
- Record: found
- Abstract: found
- Article: not found
Identification of severe acute respiratory syndrome in Canada.
- Record: found
- Abstract: found
- Article: not found
