- Record: found
- Abstract: found
- Article: found
A Toolbox for Quantitative Gene Expression in Varroa destructor: RNA Degradation in Field Samples and Systematic Analysis of Reference Gene Stability
Read this article at
Abstract
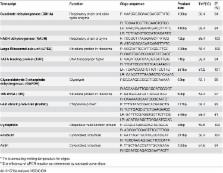
Varroa destructor is the major pest of Apis mellifera and contributes to the global honey bee health crisis threatening food security. Developing new control strategies to combat Varroa will require the application of molecular biology, including gene expression studies by quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR). Both high quality RNA samples and suitable stable internal reference genes are required for accurate gene expression studies. In this study, ten candidate genes (succinate dehydrogenase (SDHA), NADH dehydrogenase (NADH), large ribsosmal subunit, TATA-binding protein, glyceraldehyde-3-phosphate dehydrogenase, 18S rRNA (18S), heat-shock protein 90 (HSP90), cyclophilin, α-tubulin, actin), were evaluated for their suitability as normalization genes using the geNorm, Normfinder, BestKeeper, and comparative ΔCq algorithims. Our study proposes the use of no more than two of the four most stable reference genes (NADH, 18S, SDHA and HSP90) in Varroa gene expression studies. These four genes remain stable in phoretic and reproductive stage Varroa and are unaffected by Deformed wing virus load. When used for determining changes in vitellogenin gene expression, the signal-to-noise ratio (SNR) for the relatively unstable genes actin and α-tubulin was much lower than for the stable gene combinations (NADH + HSP90 +18S; NADH + HSP90; or NADH). Using both electropherograms and RT-qPCR for short and long amplicons as quality controls, we demonstrate that high quality RNA can be recovered from Varroa up to 10 days later stored at ambient temperature if collected into RNAlater and provided the body is pierced. This protocol allows the exchange of Varroa samples between international collaborators and field sample collectors without requiring frozen collection or shipping. Our results make important contributions to gene expression studies in Varroa by proposing a validated sampling protocol to obtain high quality Varroa RNA and the validation of suitable reference genes for expression studies in this globally important pest.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
Housekeeping genes as internal standards: use and limits.
- Record: found
- Abstract: found
- Article: not found
How to do successful gene expression analysis using real-time PCR.
- Record: found
- Abstract: found
- Article: not found