- Record: found
- Abstract: found
- Article: found
Convergent evolution of [ D-Leucine 1] microcystin-LR in taxonomically disparate cyanobacteria
Read this article at
Abstract
Background
Many important toxins and antibiotics are produced by non-ribosomal biosynthetic pathways. Microcystins are a chemically diverse family of potent peptide toxins and the end-products of a hybrid NRPS and PKS secondary metabolic pathway. They are produced by a variety of cyanobacteria and are responsible for the poisoning of humans as well as the deaths of wild and domestic animals around the world. The chemical diversity of the microcystin family is attributed to a number of genetic events that have resulted in the diversification of the pathway for microcystin assembly.
Results
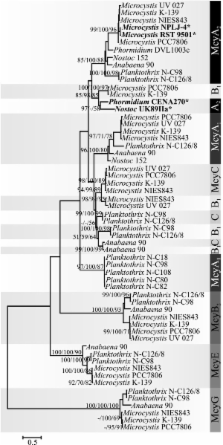
Here, we show that independent evolutionary events affecting the substrate specificity of the microcystin biosynthetic pathway have resulted in convergence on a rare [ D-Leu 1] microcystin-LR chemical variant. We detected this rare microcystin variant from strains of the distantly related genera Microcystis, Nostoc, and Phormidium. Phylogenetic analysis performed using sequences of the catalytic domains within the mcy gene cluster demonstrated a clear recombination pattern in the adenylation domain phylogenetic tree. We found evidence for conversion of the gene encoding the McyA 2 adenylation domain in strains of the genera Nostoc and Phormidium. However, point mutations affecting the substrate-binding sequence motifs of the McyA 2 adenylation domain were associated with the change in substrate specificity in two strains of Microcystis. In addition to the main [ D-Leu 1] microcystin-LR variant, these two strains produced a new microcystin that was identified as [Met 1] microcystin-LR.
Conclusions
Phylogenetic analysis demonstrated that both point mutations and gene conversion result in functional mcy gene clusters that produce the same rare [ D-Leu 1] variant of microcystin in strains of the genera Microcystis, Nostoc, and Phormidium. Engineering pathways to produce recombinant non-ribosomal peptides could provide new natural products or increase the activity of known compounds. Our results suggest that the replacement of entire adenylation domains could be a more successful strategy to obtain higher specificity in the modification of the non-ribosomal peptides than point mutations.
Related collections
Most cited references50
- Record: found
- Abstract: found
- Article: not found
A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints.
- Record: found
- Abstract: found
- Article: not found
An exact nonparametric method for inferring mosaic structure in sequence triplets.
- Record: found
- Abstract: found
- Article: not found