- Record: found
- Abstract: found
- Article: not found
COVID-19 infection in kidney transplant recipients

Read this article at
Abstract
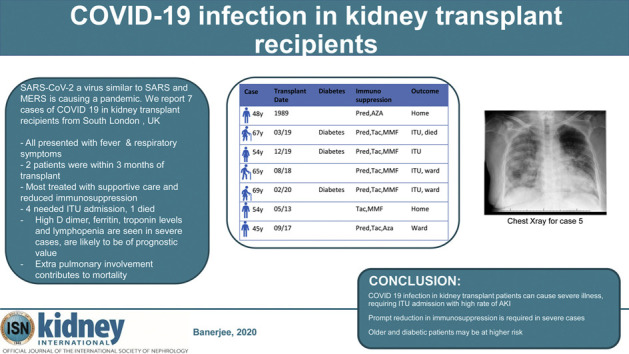
By 21 March 2020 infections related to the novel coronavirus SARS-CoV-2 had affected people from 177 countries and caused 11,252 reported deaths worldwide. Little is known about risk, presentation and outcomes of SARS-CoV-2 (COVID-19) infection in kidney transplantation recipients, who may be at high-risk due to long-term immunosuppression, comorbidity and residual chronic kidney disease. Whilst COVID-19 is predominantly a respiratory disease, in severe cases it can cause kidney and multi-organ failure. It is unknown if immunocompromised hosts are at higher risk of more severe systemic disease. Therefore, we report on seven cases of COVID-19 in kidney transplant recipients (median age 54 (range 45-69), three females, from a cohort of 2082 managed transplant follow-up patients) over a six-week period in three south London hospitals. Two of 32 patients presented within three months of transplantation. Overall, two were managed on an out-patient basis, but the remaining five required hospital admission, four in intensive care units. All patients displayed respiratory symptoms and fever. Other common clinical features included hypoxia, chest crepitation, lymphopenia and high C-reactive protein. Very high D dimer, ferritin and troponin levels occurred in severe cases and likely prognostic. Immunosuppression was modified in six of seven patients. Three patients with severe disease were diabetic. During a three week follow up one patient recovered, and one patient died. Thus, our findings suggest COVID-19 infection in kidney transplant patients may be severe, requiring intensive care admission. The symptoms are predominantly respiratory and associated with fever. Most patients had their immunosuppression reduced and were treated with supportive therapy
Graphical abstract
Related collections
Most cited references14
- Record: found
- Abstract: found
- Article: not found
Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study
- Record: found
- Abstract: found
- Article: not found
Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding

- Record: found
- Abstract: found
- Article: found