- Record: found
- Abstract: found
- Article: found
Potential ferroptosis key genes in calcific aortic valve disease
Read this article at
Abstract
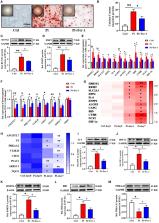
Calcific aortic valve disease (CAVD) is a highly prevalent condition that comprises a disease continuum, ranging from microscopic changes to profound fibro-calcific leaflet remodeling, culminating in aortic stenosis, heart failure, and ultimately premature death. Ferroptosis has been hypothesized to contribute to the pathogenesis of CAVD. We aimed to study the association between ferroptosis genes and CAVD and reveal the potential roles of ferroptosis in CAVD. CAVD-related differentially expressed genes (DEGs) were identified via bioinformatic analysis of Datasets GSE51472 and GSE12644 obtained from Gene Expression Omnibus. A ferroptosis dataset containing 259 genes was obtained from the Ferroptosis Database. We then intersected with CAVD-related DEGs to identify the ferroptosis DEGs. Subsequently, protein–protein interaction networks and functional enrichment analyses were performed for ferroptosis DEGs. Then, we used miRWalk3.0 to predict the target pivotal microRNAs. An in vitro model of CAVD was constructed using human aortic valve interstitial cells. The qRT-PCR and western blotting methods were used to validate the ferroptosis DEGs identified by the microarray data. A total of 21 ferroptosis DEGs in CAVD containing 12 upregulated and nine downregulated genes were identified. The results of the Gene Set Enrichment Analysis (GSEA) and analysis of the KEGG pathway by WebGestalt indicated that the ferroptosis DEGs were enriched in six signaling pathways among which NAFLD (including IL-6, BID, and PRKAA2 genes) and HIF-1 (including IL-6, HIF-1, and HMOX1 genes) signaling pathways were also verified by DAVID and/or Metascape. Finally, the in vitro results showed that the mRNA and protein expression levels of IL-6, HIF-1α, HMOX1, and BID were higher, while the levels of PRKAA2 were lower in the Pi-treated group than those in the control group. However, the addition of ferrostatin-1 (a selective ferroptosis inhibitor) significantly reversed the above changes. Therefore, IL-6, HIF-1α, HMOX1, BID, and PRKAA2 are potential key genes closely associated with ferroptosis in CAVD. Further work is required to explore the underlying ferroptosis-related molecular mechanisms and provide possible therapeutic targets for CAVD.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles
- Record: found
- Abstract: found
- Article: not found
Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease

- Record: found
- Abstract: found
- Article: found