- Record: found
- Abstract: found
- Article: found
Virulence of Vibrio alginolyticus Accentuates Apoptosis and Immune Rigor in the Oyster Crassostrea hongkongensis
Read this article at
Abstract
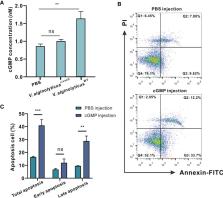
Vibrio species are ubiquitously distributed in marine environments, with important implications for emerging infectious diseases. However, relatively little is known about defensive strategies deployed by hosts against Vibrio pathogens of distinct virulence traits. Being an ecologically relevant host, the oyster Crassostrea hongkongensis can serve as an excellent model for elucidating mechanisms underlying host- Vibrio interactions. We generated a Vibrio alginolyticus mutant strain ( V. alginolyticus △ vscC ) with attenuated virulence by knocking out the vscC encoding gene, a core component of type III secretion system (T3SS), which led to starkly reduced apoptotic rates in hemocyte hosts compared to the V. alginolyticus WT control. In comparative proteomics, it was revealed that distinct immune responses arose upon encounter with V. alginolyticus strains of different virulence. Quite strikingly, the peroxisomal and apoptotic pathways are activated by V. alginolyticus WT infection, whereas phagocytosis and cell adhesion were enhanced in V. alginolyticus △ vscC infection. Results for functional studies further show that V. alginolyticus WT strain stimulated respiratory bursts to produce excess superoxide (O2 •−) and hydrogen peroxide (H 2O 2) in oysters, which induced apoptosis regulated by p53 target protein (p53tp). Simultaneously, a drop in sGC content balanced off cGMP accumulation in hemocytes and repressed the occurrence of apoptosis to a certain extent during V. alginolyticus △ vscC infection. We have thus provided the first direct evidence for a mechanistic link between virulence of Vibrio spp. and its immunomodulation effects on apoptosis in the oyster. Collectively, we conclude that adaptive responses in host defenses are partially determined by pathogen virulence, in order to safeguard efficiency and timeliness in bacterial clearance.
Related collections
Most cited references94
- Record: found
- Abstract: found
- Article: not found
Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.
- Record: found
- Abstract: found
- Article: not found
TBtools - an integrative toolkit developed for interactive analyses of big biological data

- Record: found
- Abstract: found
- Article: found