- Record: found
- Abstract: found
- Article: found
The compensatory potential of increased immigration following intensive American mink population control is diluted by male-biased dispersal
Read this article at
Abstract
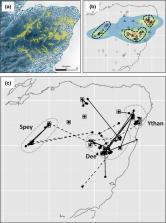
Attempts to mitigate the impact of invasive species on native ecosystems increasingly target large land masses where control, rather than eradication, is the management objective. Depressing numbers of invasive species to a level where their impact on native biodiversity is tolerable requires overcoming the impact of compensatory immigration from non-controlled portions of the landscape. Because of the expected scale-dependency of dispersal, the overall size of invasive species management areas relative to the dispersal ability of the controlled species will determine the size of any effectively conserved core area unaffected by immigration from surrounding areas. However, when dispersal is male-biased, as in many mammalian invasive carnivores, males may be overrepresented amongst immigrants, reducing the potential growth rate of invasive species populations in re-invaded areas. Using data collected from a project that gradually imposed spatially comprehensive control on invasive American mink ( Neovison vison) over a 10,000 km 2 area of NE Scotland, we show that mink captures were reduced to almost zero in 3 years, whilst there was a threefold increase in the proportion of male immigrants. Dispersal was often long distance and linking adjacent river catchments, asymptoting at 38 and 31 km for males and females respectively. Breeding and dispersal were spatially heterogeneous, with 40 % of river sections accounting for most captures of juvenile (85 %), adult female (65 %) and immigrant (57 %) mink. Concentrating control effort on such areas, so as to turn them into “attractive dispersal sinks” could make a disproportionate contribution to the management of recurrent re-invasion of mainland invasive species management areas.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: not found
COLONY: a program for parentage and sibship inference from multilocus genotype data.
- Record: found
- Abstract: not found
- Article: not found
Estimating Relatedness Using Genetic Markers

- Record: found
- Abstract: found
- Article: found