- Record: found
- Abstract: found
- Article: found
CTCF as a multifunctional protein in genome regulation and gene expression
Read this article at
Abstract
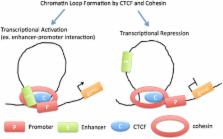
CCCTC-binding factor (CTCF) is a highly conserved zinc finger protein and is best known as a transcription factor. It can function as a transcriptional activator, a repressor or an insulator protein, blocking the communication between enhancers and promoters. CTCF can also recruit other transcription factors while bound to chromatin domain boundaries. The three-dimensional organization of the eukaryotic genome dictates its function, and CTCF serves as one of the core architectural proteins that help establish this organization. The mapping of CTCF-binding sites in diverse species has revealed that the genome is covered with CTCF-binding sites. Here we briefly describe the diverse roles of CTCF that contribute to genome organization and gene expression.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
A map of the cis-regulatory sequences in the mouse genome.
- Record: found
- Abstract: found
- Article: not found
Cohesin mediates transcriptional insulation by CCCTC-binding factor.
- Record: found
- Abstract: found
- Article: not found