- Record: found
- Abstract: found
- Article: found
Epigenetic features of FoxP3 in children with cow’s milk allergy
Read this article at
Abstract
Background
DNA methylation of the Th1 and Th2 cytokine genes is altered during cow’s milk allergy (CMA). Forkhead box transcription factor 3 ( FoxP3) is essential for the development and function of regulatory T cells (Tregs) and is involved in oral tolerance acquisition. We assessed whether tolerance acquisition in children with IgE-mediated CMA is associated with DNA demethylation of the Treg-specific demethylated region (TSDR) of FoxP3.
Results
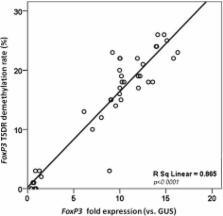
Forty children (aged 3–18 months) were enrolled: 10 children with active IgE-mediated CMA (group 1), 10 children who outgrew CMA after dietary treatment with an extensively hydrolyzed casein formula containing the probiotic Lactobacillus rhamnosus GG (group 2), 10 children who outgrew CMA after treatment with other formulas (group 3), and 10 healthy controls (group 4). FoxP3 TSDR demethylation and expression were measured in mononuclear cells purified from peripheral blood of the four groups of children. FoxP3 TSDR demethylation was significantly lower in children with active IgE-mediated CMA than in either children who outgrew CMA or in healthy children. Formula selection influenced the FoxP3 TSDR demethylation profile. The FoxP3 TSDR demethylation rate and expression level were correlated.
Related collections
Most cited references16
- Record: found
- Abstract: found
- Article: not found
DNA methylation profiling of human chromosomes 6, 20 and 22

- Record: found
- Abstract: found
- Article: found
Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants
- Record: found
- Abstract: found
- Article: not found