- Record: found
- Abstract: found
- Article: found
The life cycle of a genome project: perspectives and guidelines inspired by insect genome projects
Read this article at
Abstract
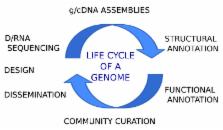
Many research programs on non-model species biology have been empowered by genomics. In turn, genomics is underpinned by a reference sequence and ancillary information created by so-called “genome projects”. The most reliable genome projects are the ones created as part of an active research program and designed to address specific questions but their life extends past publication. In this opinion paper I outline four key insights that have facilitated maintaining genomic communities: the key role of computational capability, the iterative process of building genomic resources, the value of community participation and the importance of manual curation. Taken together, these ideas can and do ensure the longevity of genome projects and the growing non-model species community can use them to focus a discussion with regards to its future genomic infrastructure.
Related collections
Most cited references21
- Record: found
- Abstract: not found
- Article: not found
The Human Genome Project: lessons from large-scale biology.

- Record: found
- Abstract: found
- Article: found