- Record: found
- Abstract: found
- Article: found
The DDN Catalytic Motif Is Required for Metnase Functions in Non-homologous End Joining (NHEJ) Repair and Replication Restart*
Read this article at
Abstract
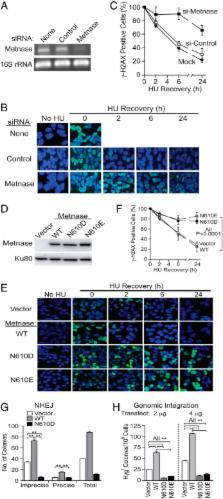
Background: Metnase, a transposase-containing DNA repair protein, retains DNA cleavage activity with a DDN motif.
Results: Substitution with the ancestral transposase DDD/DDE catalytic motif results in a decrease in ssDNA binding and ss-overhang cleavage activities.
Conclusion: The DDN motif is required for Metnase DNA repair activities.
Significance: Understanding the requirements for catalytic activity provides insights on how Metnase functions as a DNA repair protein.
Abstract
Metnase (or SETMAR) arose from a chimeric fusion of the Hsmar1 transposase downstream of a protein methylase in anthropoid primates. Although the Metnase transposase domain has been largely conserved, its catalytic motif (DDN) differs from the DDD motif of related transposases, which may be important for its role as a DNA repair factor and its enzymatic activities. Here, we show that substitution of DDN 610 with either DDD 610 or DDE 610 significantly reduced in vivo functions of Metnase in NHEJ repair and accelerated restart of replication forks. We next tested whether the DDD or DDE mutants cleave single-strand extensions and flaps in partial duplex DNA and pseudo-Tyr structures that mimic stalled replication forks. Neither substrate is cleaved by the DDD or DDE mutant, under the conditions where wild-type Metnase effectively cleaves ssDNA overhangs. We then characterized the ssDNA-binding activity of the Metnase transposase domain and found that the catalytic domain binds ssDNA but not dsDNA, whereas dsDNA binding activity resides in the helix-turn-helix DNA binding domain. Substitution of Asn-610 with either Asp or Glu within the transposase domain significantly reduces ssDNA binding activity. Collectively, our results suggest that a single mutation DDN 610 → DDD 610, which restores the ancestral catalytic site, results in loss of function in Metnase.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast.
- Record: found
- Abstract: found
- Article: not found
Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system.
- Record: found
- Abstract: found
- Article: not found