- Record: found
- Abstract: found
- Article: found
Gene Expression System in Green Sulfur Bacteria by Conjugative Plasmid Transfer
Read this article at
Abstract
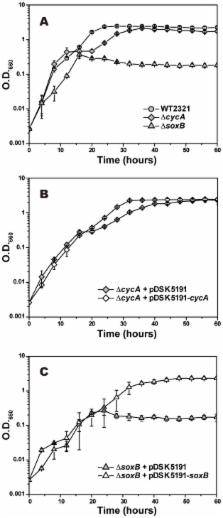
Gene transfer and expression systems in green sulfur bacteria were established by bacterial conjugation with Escherichia coli. Conjugative plasmid transfer from E. coli S17-1 to a thermophilic green sulfur bacterium, Chlorobaculum tepidum (formerly Chlorobium tepidum) WT2321, was executed with RSF1010-derivative broad-host-range plasmids, named pDSK5191 and pDSK5192, that confer erythromycin and streptomycin/spectinomycin resistance, respectively. The transconjugants harboring these plasmids were reproducibly obtained at a frequency of approximately 10 -5 by selection with erythromycin and a combination of streptomycin and spectinomycin, respectively. These plasmids were stably maintained in C. tepidum cells in the presence of these antibiotics. The plasmid transfer to another mesophilic green sulfur bacterium, C. limnaeum (formerly Chlorobium phaeobacteroides) RK-j-1, was also achieved with pDSK5192. The expression plasmid based on pDSK5191 was constructed by incorporating the upstream and downstream regions of the pscAB gene cluster on the C. tepidum genome, since these regions were considered to include a constitutive promoter and a ρ-independent terminator, respectively. Growth defections of the ∆ cycA and ∆ soxB mutants were completely rescued after introduction of pDSK5191- cycA and - soxB that were designed to express their complementary genes. On the other hand, pDSK5191- 6xhis-pscAB, which incorporated the gene cluster of 6xhis-pscA and pscB, produced approximately four times more of the photosynthetic reaction center complex with His-tagged PscA as compared with that expressed in the genome by the conventional natural transformation method. This expression system, based on conjugative plasmid, would be applicable to general molecular biological studies of green sulfur bacteria.
Related collections
Most cited references34
- Record: found
- Abstract: not found
- Article: not found
GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes.
- Record: found
- Abstract: found
- Article: not found