- Record: found
- Abstract: found
- Article: not found
Klebsazolicin inhibits 70S ribosome by obstruction of the peptide exit tunnel
Abstract
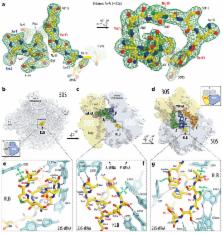
While screening of small-molecular metabolites produced by most cultivatable microorganisms often results in rediscovery of known compounds, genome-mining programs allow to harness much greater chemical diversity and result in discovery of new molecular scaffolds. Here we report genome-guided identification of a new antibiotic klebsazolicin (KLB) from Klebsiella pneumoniae that inhibits growth of sensitive cells by targeting ribosome. A member of ribosomally-synthesized post-translationally modified peptides (RiPPs), KLB is characterized by the presence of unique N-terminal amidine ring essential for its activity. Biochemical in vitro studies indicate that KLB inhibits ribosome by interfering with translation elongation. Structural analysis of the ribosome-KLB complex reveals the compound bound in the peptide exit tunnel overlapping with the binding sites of macrolides or streptogramins-B. KLB adopts compact conformation and largely obstructs the tunnel. Engineered KLB fragments retain in vitro activity and can serve as a starting point for the development of new bioactive compounds.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
Antibiotic resistance-the need for global solutions.
- Record: found
- Abstract: found
- Article: not found
Human commensals producing a novel antibiotic impair pathogen colonization.
- Record: found
- Abstract: found
- Article: not found
