- Record: found
- Abstract: found
- Article: found
Antennal transcriptomic analysis of carboxylesterases and glutathione S-transferases associated with odorant degradation in the tea gray geometrid, Ectropis grisescens (Lepidoptera, Geometridae)
Read this article at
Abstract
Introduction: Carboxylesterases (CXEs) and glutathione S-transferases (GSTs) can terminate olfactory signals during chemosensation by rapid degradation of odorants in the vicinity of receptors. The tea grey geometrid, Ectropis grisescens (Lepidoptera, Geometridae), one of the most devastating insect herbivores of tea plants in China, relies heavily on plant volatiles to locate the host plants as well as the oviposition sites. However, CXEs and GSTs involved in signal termination and odorant clearance in E. grisescens remains unknown.
Methods: In this study, identification and spatial expression profiles of CXEs and GSTs in this major tea pest were investigated by transcriptomics and qRT-PCR, respectively.
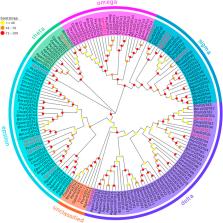
Results: As a result, we identified 28 CXEs and 16 GSTs from female and male antennal transcriptomes. Phylogenetic analyses clustered these candidates into several clades, among which antennal CXEs, mitochondrial and cytosolic CXEs, and delta group GSTs contained genes commonly associated with odorants degradation. Spatial expression profiles showed that most CXEs (26) were expressed in antennae. In comparison, putative GSTs exhibited a diverse expression pattern across different tissues, with one GST expressed specifically in the male antennae.
Disscussion: These combined results suggest that 12 CXEs (EgriCXE1, 2, 4, 6, 8, 18, 20-22, 24, 26, and 29) and 5 GSTs (EgriGST1 and EgriGST delta group) provide a major source of candidate genes for odorants degradation in E. grisescens.
Related collections
Most cited references82
- Record: found
- Abstract: found
- Article: not found
Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.
- Record: found
- Abstract: found
- Article: not found
MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms.
- Record: found
- Abstract: found
- Article: not found