- Record: found
- Abstract: found
- Article: found
Comparative mode of action of the antimicrobial peptide melimine and its derivative Mel4 against Pseudomonas aeruginosa

Read this article at
Abstract
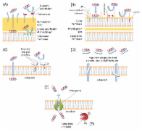
Melimine and Mel4 are chimeric cationic peptides with broad-spectrum antimicrobial activity. They have been shown to be highly biocompatible in animal models and human clinical trials. The current study examined the mechanism of action of these two antimicrobial peptides against P. aeruginosa. The effect of the peptides of endotoxin neutralization, and their interactions with cytoplasmic membranes using DiSC(3)-5 and Sytox green, Syto-9 and PI dyes were analysed. Release of ATP and DNA/RNA were determined using ATP luminescence and increase in OD 260 nm. The bacteriolytic ability of the peptides was determined by measuring decreases in OD 620 nm. Both the peptides neutralized LPS suggesting their interaction with lipid A. Cytoplasmic membrane was disrupted within 30 seconds, which correlated with reductions in cellular viability. At 2 minutes melimine or Mel4, released 75% and 36% cellular ATP respectively (P < 0.001). Membrane permeabilization started 5 minutes with simultaneous release of DNA/RNA. Flow cytometry demonstrated 52% and 18% bacteria were stained with PI after 30 minutes. Overall, melimine showed higher capacity for membrane disruption compared to Mel4 (P < 0.001). The findings of this study have been summarized as a timeline of bactericidal activity, suggesting that the peptides permeabilized P. aeruginosa within 5 minutes, started lysis within 2 hours of exposure.
Related collections
Most cited references52
- Record: found
- Abstract: found
- Article: not found