- Record: found
- Abstract: found
- Article: not found
Host-membrane interacting interface of the SARS coronavirus envelope protein: Immense functional potential of C-terminal domain
Read this article at
Abstract
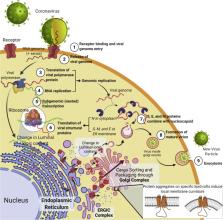
The Envelope (E) protein in SARS Coronavirus (CoV) is a small structural protein, incorporated as part of the envelope. A major fraction of the protein has been known to be associated with the host membranes, particularly organelles related to intracellular trafficking, prompting CoV packaging and propagation. Studies have elucidated the central hydrophobic transmembrane domain of the E protein being responsible for much of the viroporin activity in favor of the virus. However, newer insights into the organizational principles at the membranous compartments within the host cells suggest further complexity of the system. The lesser hydrophobic Carboxylic-terminal of the protein harbors interesting amino acid sequences- suggesting at the prevalence of membrane-directed amyloidogenic properties that remains mostly elusive. These highly conserved segments indicate at several potential membrane-associated functional roles that can redefine our comprehensive understanding of the protein. This should prompt further studies in designing and characterizing of effective targeted therapeutic measures.
Graphical abstract
Highlights
-
•
The SARS CoV Envelope protein is a small structural protein of the virus that has been suggested to have significant viroporin like activity.
-
•
Majority of its function is mediated at the interface of host-membrane interactions.
-
•
Focus at the membrane-directed features of the protein provide useful insight into gaining mechanistic insight into its viroporin functions.
-
•
Studies have elaborated the central hydrophobic transmembrane domain of E protein, known to affect ion-channel formation.
-
•
The C-terminal region of the protein show further potential host-membrane directed functional roles.
-
•
The highly conserved amyloidogenic amino acid stretches of the C-terminal suggest at significant contribution to CoV propagation.
-
•
Comprehensive understanding of the roles of each segment of the protein can help in defining newer therapeutic target sequences.
Related collections
Most cited references91
- Record: found
- Abstract: found
- Article: not found
Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding
- Record: found
- Abstract: found
- Article: not found
Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia.
- Record: found
- Abstract: found
- Article: found

