- Record: found
- Abstract: found
- Article: found
Diffusion, Crowding & Protein Stability in a Dynamic Molecular Model of the Bacterial Cytoplasm
Read this article at
Abstract
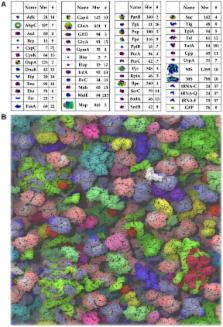
A longstanding question in molecular biology is the extent to which the behavior of macromolecules observed in vitro accurately reflects their behavior in vivo. A number of sophisticated experimental techniques now allow the behavior of individual types of macromolecule to be studied directly in vivo; none, however, allow a wide range of molecule types to be observed simultaneously. In order to tackle this issue we have adopted a computational perspective, and, having selected the model prokaryote Escherichia coli as a test system, have assembled an atomically detailed model of its cytoplasmic environment that includes 50 of the most abundant types of macromolecules at experimentally measured concentrations. Brownian dynamics (BD) simulations of the cytoplasm model have been calibrated to reproduce the translational diffusion coefficients of Green Fluorescent Protein (GFP) observed in vivo, and “snapshots” of the simulation trajectories have been used to compute the cytoplasm's effects on the thermodynamics of protein folding, association and aggregation events. The simulation model successfully describes the relative thermodynamic stabilities of proteins measured in E. coli, and shows that effects additional to the commonly cited “crowding” effect must be included in attempts to understand macromolecular behavior in vivo.
Author Summary
The interior of a typical bacterial cell is a highly crowded place in which molecules must jostle and compete with each other in order to carry out their biological functions. The conditions under which such molecules are typically studied in vitro, however, are usually quite different: one or a few different types of molecules are studied as they freely diffuse in a dilute, aqueous solution. There is therefore a significant disconnect between the conditions under which molecules can be most usefully studied and the conditions under which such molecules usually “live”, and developing ways to bridge this gap is likely to be important for properly understanding molecular behavior in vivo. Toward this end, we show in this work that computer simulations can be used to model the interior of bacterial cells at a near atomic level of detail: the rates of diffusion of proteins are matched to known experimental values, and their thermodynamic stabilities are found to be in good agreement with the few measurements that have so far been performed in vivo. While the simulation approach is certainly not free of assumptions, it offers a potentially important complement to experimental techniques and provides a vivid illustration of molecular behavior inside a biological cell that is likely to be of significant educational value.
Related collections
Most cited references68
- Record: found
- Abstract: found
- Article: not found
Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation.
- Record: found
- Abstract: found
- Article: not found
A graph-theory algorithm for rapid protein side-chain prediction.
- Record: found
- Abstract: found
- Article: not found