- Record: found
- Abstract: found
- Article: found
Wide spectrum and high frequency of genomic structural variation, including transposable elements, in large double-stranded DNA viruses
Read this article at
Abstract
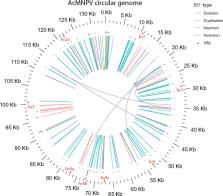
Our knowledge of the diversity and frequency of genomic structural variation segregating in populations of large double-stranded (ds) DNA viruses is limited. Here, we sequenced the genome of a baculovirus ( Autographa californica multiple nucleopolyhedrovirus [AcMNPV]) purified from beet armyworm ( Spodoptera exigua) larvae at depths >195,000× using both short- (Illumina) and long-read (PacBio) technologies. Using a pipeline relying on hierarchical clustering of structural variants (SVs) detected in individual short- and long-reads by six variant callers, we identified a total of 1,141 SVs in AcMNPV, including 464 deletions, 443 inversions, 160 duplications, and 74 insertions. These variants are considered robust and unlikely to result from technical artifacts because they were independently detected in at least three long reads as well as at least three short reads. SVs are distributed along the entire AcMNPV genome and may involve large genomic regions (30,496 bp on average). We show that no less than 39.9 per cent of genomes carry at least one SV in AcMNPV populations, that the vast majority of SVs (75%) segregate at very low frequency (<0.01%) and that very few SVs persist after ten replication cycles, consistent with a negative impact of most SVs on AcMNPV fitness. Using short-read sequencing datasets, we then show that populations of two iridoviruses and one herpesvirus are also full of SVs, as they contain between 426 and 1,102 SVs carried by 52.4–80.1 per cent of genomes. Finally, AcMNPV long reads allowed us to identify 1,757 transposable elements (TEs) insertions, 895 of which are truncated and occur at one extremity of the reads. This further supports the role of baculoviruses as possible vectors of horizontal transfer of TEs. Altogether, we found that SVs, which evolve mostly under rapid dynamics of gain and loss in viral populations, represent an important feature in the biology of large dsDNA viruses.
Related collections
Most cited references75
- Record: found
- Abstract: found
- Article: not found
Mutational and fitness landscapes of an RNA virus revealed through population sequencing.
- Record: found
- Abstract: not found
- Article: not found